Non-canonical DNA structures found in the control centre for the activation of HIV-1 genes
|After a long and intense competition, the structure of the DNA was described for the first time in 1953 by James Watson, Francis Crick, Rosalind Franklin, and Maurice Wilkins. However, the secrets of the DNA are still unfolding in front of our eyes. Other than the iconic double helix, the DNA can also adopt alternative structures depending on conditions such as hydration, temperature, pH, and its primary sequence. In 2018 the i-motifs were discovered in cellular DNA; they are a four-strand DNA structure that can be found in regulatory regions where there is an abundance of cytosines, one of the four letters of the DNA alphabet. These structures were found to be involved in the regulation and functioning of the genes.
Since then, several groups around the world have focused on the presence and role of i-motifs in human cells. In 2019, a research team led by Sara Richter, Professor at University of Padova and Janez Plavec, director of the Slovenian CERIC Partner Facility at the National Institute of Chemistry, highlighted the presence of this non-canonical DNA structure in viral DNA. The work was performed with the major contribution of Dr. Emanuela Ruggiero. For the first time, the i-motifs were found in the genome of HIV-1 virus, again in a regulatory region. Several instruments and techniques were employed to analyse and confirm the presence of i-motifs, and namely 600 MHz Nuclear Magnetic Resonance at the SloNMR centre in Ljubljana.
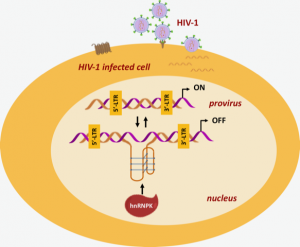
The region where the i-motif was found represents the control centre for the activation of HIV genes. The authors propose that these i-motifs are triggered during the infection to modulate viral processes. Moreover, the scientists demonstrated that a specific i-motif interacts with the human ribonucleoprotein (hnRNP) K, a multifunctional protein involved in the regulation of various fundamental processes of human cells. The results of this research, which links the presence of i-motifs in the HIV-1 genome and their interaction with cellular proteins, not only provide valuable insights into the complexity of the HIV-1 genome, but also lay the necessary information for a possible innovative antiviral drug design.



