Molecular mechanism potentially involved in gene regulation crucial for brain development revealed
|Several diseases are associated with the expansion of short nucleotide repeats, in the form of non-canonical structures (such as hairpins, i-motifs or G-quadruplexes) dispersed across genes. In this context, a CGAG-rich region present in the Autism Susceptibility 2 (AUTS2) promoter gene appears to be involved in the switch between two isoforms (full-length and C-terminal) of the encoded protein, which plays an important role in brain development and neuronal differentiation.
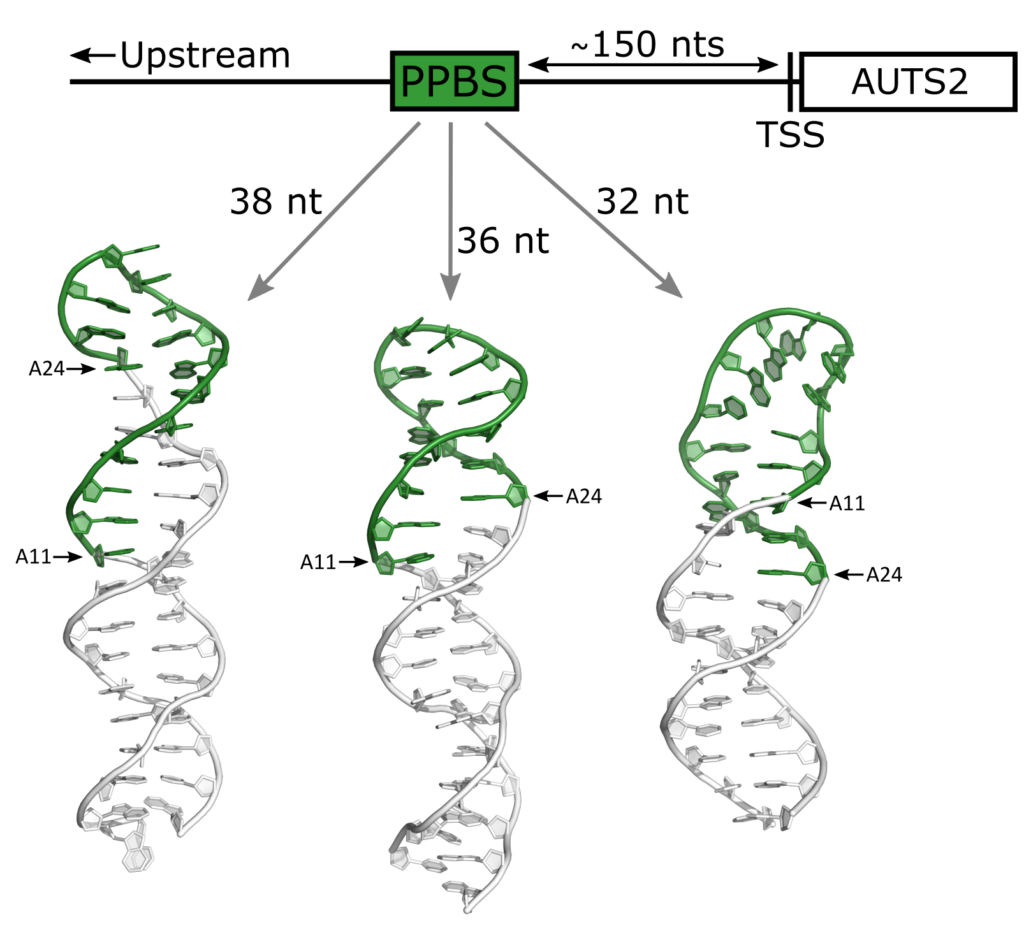
To better understand the molecular mechanism responsible for this switch, Dr. Aleš Novotný, Prof. Janez Plavec and Dr. Vojč Kocman of the Slovenian NMR Centre (Ljubljana) studied a 60 nt long CGAG-rich region of the AUTS2 gene, that can adopt a variety of non-canonical secondary structures. However, due to the flexibility and polymorphism expressed by this region, it was hard to structurally characterise it. Hence, using the Nuclear Magnetic Resonance Spectrometers DAVID and LARA of the CERIC Slovenian partner facility at the National Institute of Chemistry, researchers focused on three shorter oligonucleotides containing CGAG repeats, which surround the putative protein binding site for transcription regulatory proteins (located in the loop of the CGAG-region).
Scientists then discovered that these short sequences adopt thermally stable non-canonical hairpin structures stabilized by repeating structural motifs called CGAG blocks. These motifs are formed consecutively, in a way that exploits a shift in register to have the maximum of consecutive G:C and G:A base pairs. The differences in CGAG repeat shifting affect the structure of the loop region, leading to different folds in the CGAG-rich region, change in the protein binding site accessibility, and potentially causing the switch between the full-length and C-terminal AUTS2 isoforms.
ORIGINAL ARTICLE:



