New insights on the formation of secondary nucleic acid structures adopted by cytosine-rich DNA
|i-Motifs (iMs) are tetra-helical secondary structures, generated by the folding of DNA sequences containing at least four runs of repetitive cytosines, that could have important therapeutical applications (due to their activity as gene promoters and oncogene regulators) and support technologically innovative approaches (such as bio-sensing and drug delivery). However, until recent days there was a poor understanding of which were the conditions needed for DNA sequences to fold, and there wasn’t any algorithm able to predict iM folding from the primary nucleotide sequence.
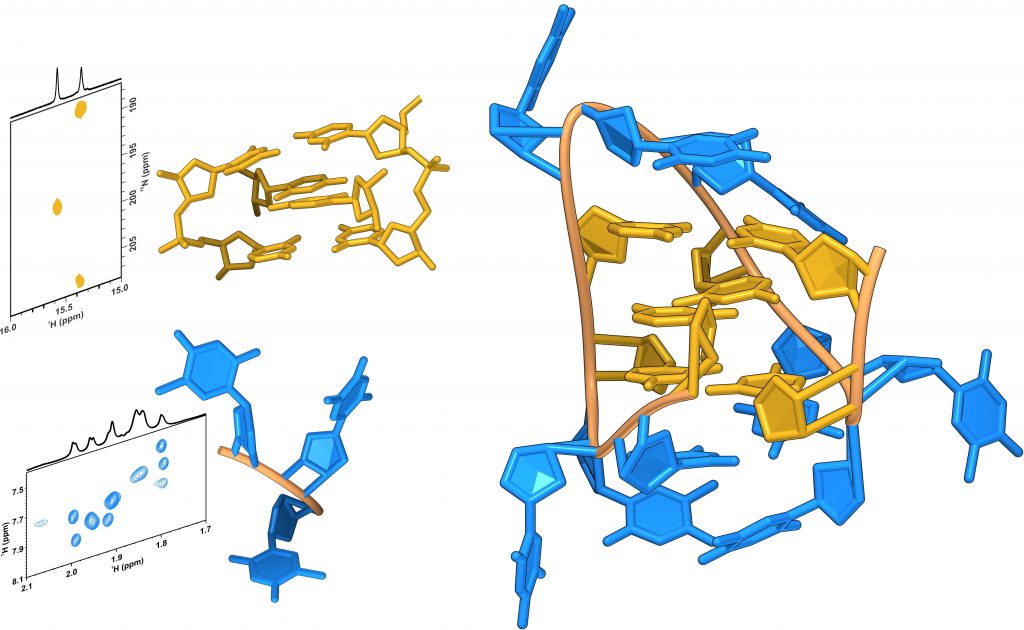
Then, Dr. Michele Ghezzo and Prof. Claudia Sissi (University of Padua), together with Dr. Marko Trajkovski and Prof. Janez Plavec (Slovenian NMR Centre, National Institute of Chemistry), developed a novel pipeline for the systematic screening of iM-forming model sequences, in order to provide a description of this process at the molecular level and understand what is the minimal length of the loops required for formation of an intra-molecular iM. At each step of this protocol, the folding of the selected sequence into a 3 CC+ base-paired intra-molecular iM was assessed by nuclear magnetic resonance spectroscopy (600 and 800 MHz NMR spectrometers, available at the Slovenian CERIC partner facility at the National Institute of Chemistry in Ljubljana), and other techniques.
Scientists discovered that two and three nucleotides are required to connect the strands through the minor and major grooves of the iM, respectively, and that there is an asymmetric behavior according to the distribution of the cytosines. Hopefully, these results could be helpful to develop prediction tools for the identification of biologically functional iMs, and to create technological innovative devices based on these secondary structures.
ORIGINAL ARTICLE:
A Screening Protocol for Exploring Loop Length Requirements for the Formation of a Three Cytosine-Cytosine+ Base-Paired i-Motif
Ghezzo M., Trajkovski M., Plavec J., Sissi C., Angew. Chem. Int. Ed., 2023, DOI



