Study on DNA origami structures with gold nanoparticles gives new hints for nanoscale manufacturing
|Origami is the art of paper folding often associated with Japanese culture. Researchers from the Faculty of Physics and Centre for Nanoscience (CeNS) in Munich and the Institute of Inorganic Chemistry at the Graz University of Technology, conducted an experiment using origami technique, using an object profoundly different from paper: the DNA molecule.
The DNA is a molecule composed of two chains made of nucleotides, which coil around each other to form a double helix. Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine, guanine, adenine or thymine), a sugar called deoxyribose, and a phosphate group. The nitrogenous bases are bound together according to the basic coupling rules: adenine with thymine and cytosine with guanine. This property allows creating two- and three-dimensional shapes at the nanoscale as the origami with the paper.
DNA origami, in combination with other components, such as metals, has proven to be extremely effective for the design and manufacturing of nanomaterials with applications in fields from biomedicine to electronics.
In experiments led by Caroline Hartl, the teams around Tim Liedl and Bert Nickel used Small-Angle X-ray scattering (SAXS) at the Austrian CERIC Partner Facility at the Elettra synchrotron in Trieste, to investigate on how DNA origami allows accurately positioning guest molecules in three dimensions. Indeed, one of the most prominent features of DNA assemblies is the possibility to place and control guest molecules and particles with high precision.
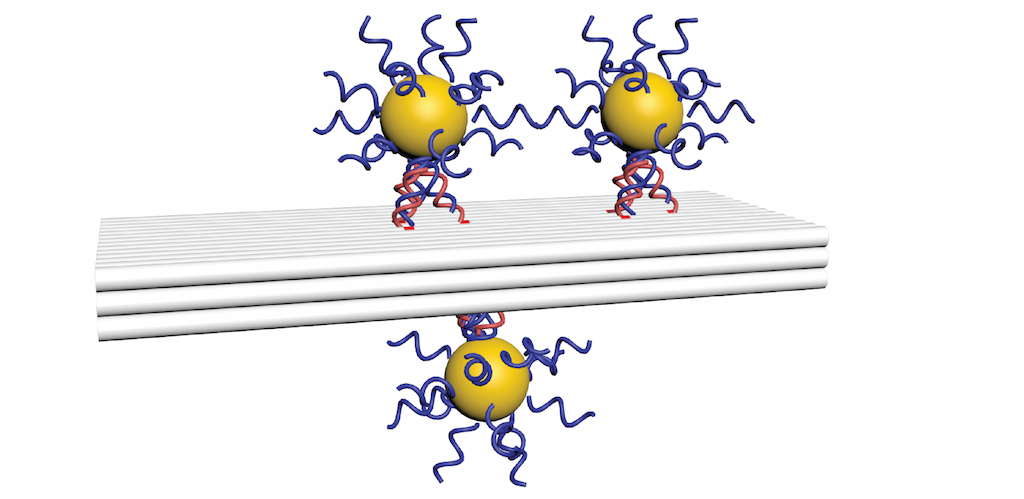 The image shows a DNA object (grey) on which we have placed gold nanoparticles. We used SAXS to triangulate the distances between the particles and test for precision of placement.
The image shows a DNA object (grey) on which we have placed gold nanoparticles. We used SAXS to triangulate the distances between the particles and test for precision of placement.
While DNA origami itself has fairly predictable spacing properties along the axis of DNA, the attachment of relatively large structures such as the Gold NanoParticles (AuNPs) of 10 nm diameter used in this experiment requires multiple attachment strands, adding further complexity. In this study, SAXS measurements provided information on the distance and disposition of AuNPs within an uncertainty range of solely 1.2 nm. The experiment demonstrated that it is possible to place metal nanoparticles in a DNA molecule with very high precision by carefully designing the AuNP attachment sites on the DNA-structure and by fine-tuning the connector types.
The results of this study give additional evidence on the effectiveness of DNA self-assembly in the spatial arrangement of metal nanoparticles into nano-antennae or more complex architectures, adding a new brick to the knowledge needed for the nanofabrication of biomedical, electronic and photonic devices and materials.



